Biology has evolved far beyond petri dishes and microscopes. Today, it thrives in the realm of data-massive, complex, and multidimensional data generated by omics technologies like genomics, transcriptomics, proteomics, metabolomics, and epigenomics. But making sense of this data tsunami requires more than biology-it demands code. Specifically, it demands R and Python, the two most powerful and accessible programming languages for modern biological data analysis.
This article explores why these languages have become the backbone of omics research, what types of analysis they support, how they differ and complement each other, and where beginners can learn them effectively-with direct links to beginner-friendly courses on the LBRN Training Platform.
Why R and Python Matter in Omics
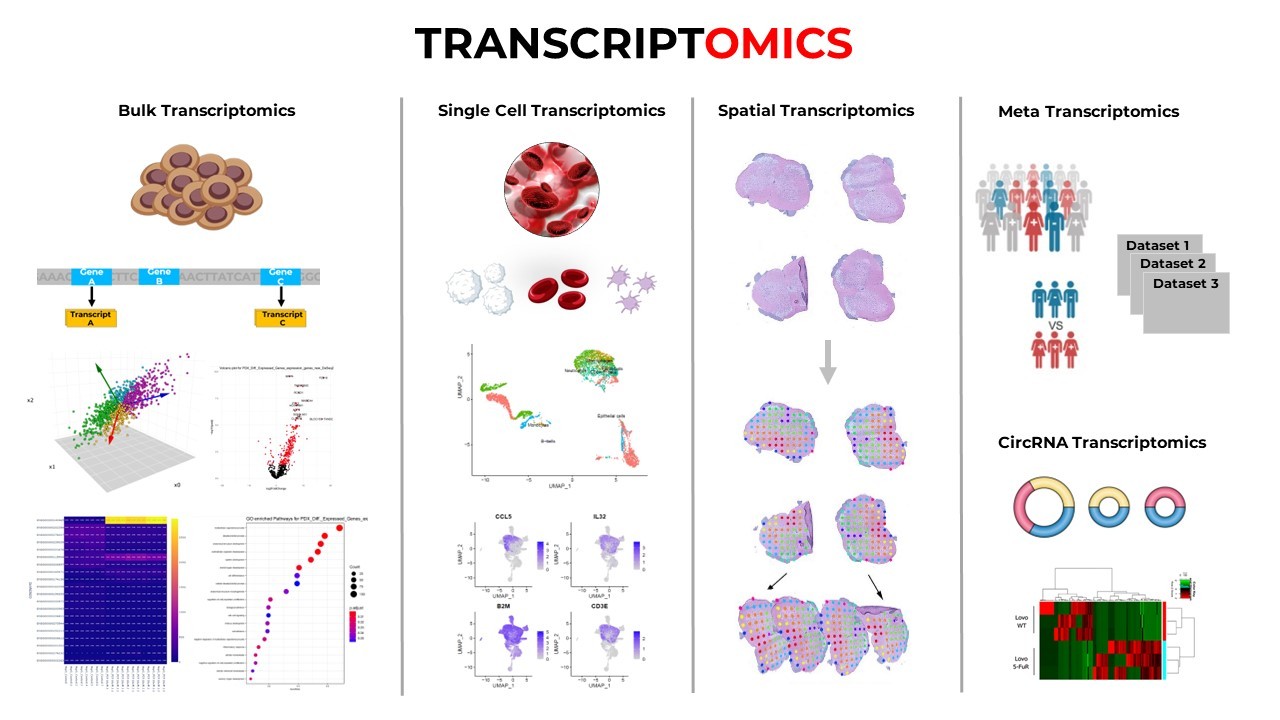
Omics datasets are vast and multidimensional. A single RNA-seq or whole-genome experiment may produce millions of reads across thousands of genes or loci. Making biological sense of such high-throughput data requires computational tools that are flexible, reproducible, and scalable-all hallmarks of R and Python.
These languages let researchers:
-
Process raw sequencing data
-
Perform quality control
-
Analyze gene expression or variation
-
Visualize complex patterns
-
Build predictive or classification models
-
Integrate multi-omics layers
They also ensure reproducibility-an essential requirement in modern computational biology-through scripting, version control, and open-source sharing.
R: The Statistical Powerhouse
R is a language built by statisticians for data analysis, which is why it has long been favored in transcriptomics and differential expression studies. With thousands of packages tailored for biological data (especially through Bioconductor), R offers specialized workflows for each omics domain.
In transcriptomics, tools like DESeq2, edgeR, and limma enable differential expression analysis. For single-cell RNA-seq, the Seurat package enables clustering, dimensionality reduction, trajectory inference, and cell type annotation.
For epigenomics, packages like minfi, ChAMP, and annotatr allow DNA methylation and histone modification analysis. In proteomics, MSnbase and DEP are used to interpret mass spectrometry data.
R’s standout feature is its visualization ecosystem. Tools like ggplot2, pheatmap, ComplexHeatmap, and plotly allow for stunning, informative visualizations essential for publications and insights.
✅ Start learning R for omics today with LBRN’s beginner-friendly course:
R for Omics Data Analysis – LBRN Training Platform
Python: The Flexible Power Engine
Python is known for its readable syntax and general-purpose capabilities. It excels in workflow automation, machine learning, and handling large-scale genomic data. Python has carved a strong niche in fields such as genomics, metagenomics, and AI-powered omics.
For genomic analysis, libraries like Biopython, pysam, and pybedtools allow easy manipulation of sequences, alignments, and variant data. In metagenomics, Python supports QIIME 2, a dominant pipeline for microbial diversity and community profiling.
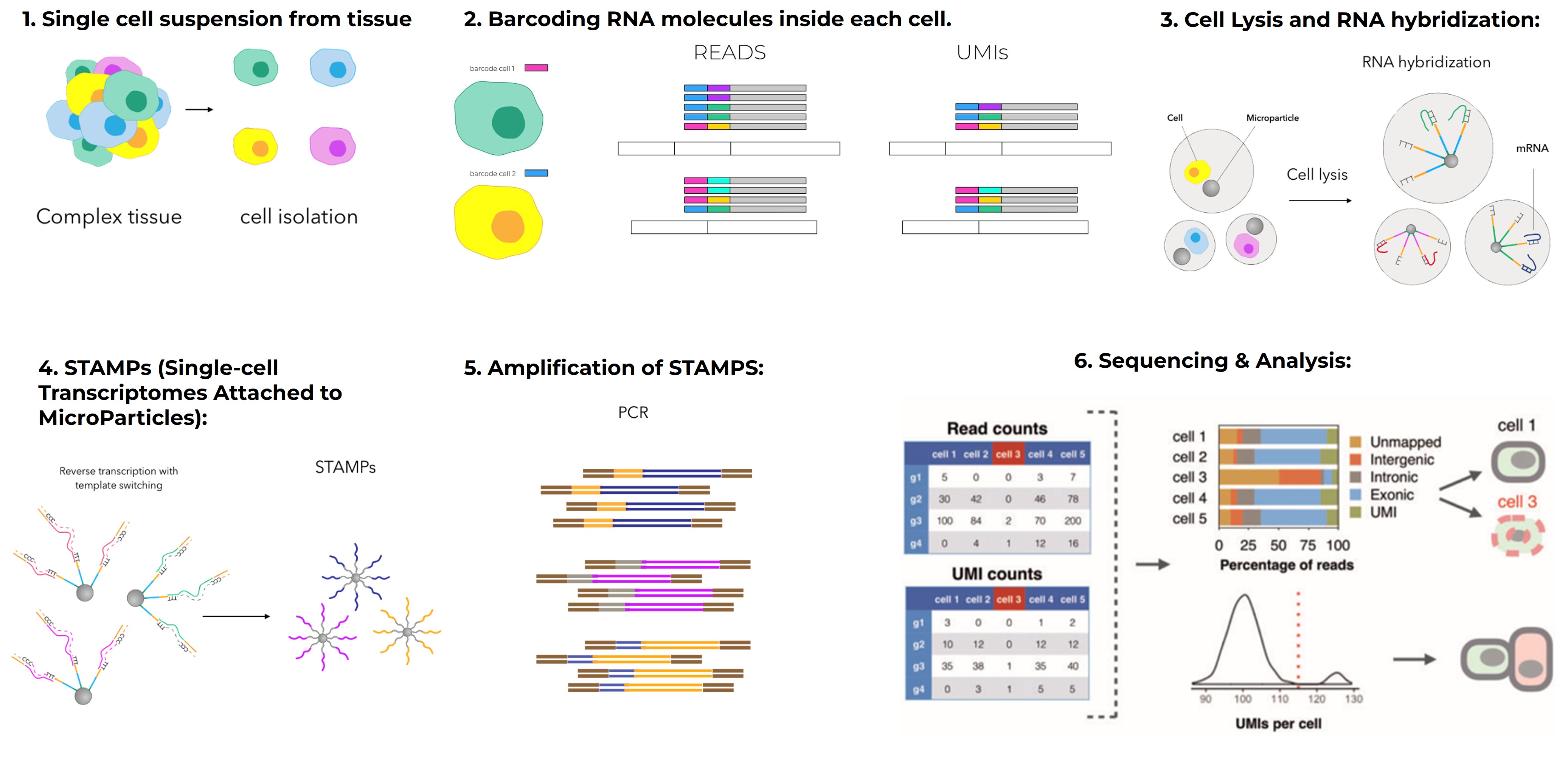
In single-cell transcriptomics, Scanpy is Python’s counterpart to Seurat. It integrates with anndata, UMAP, and matplotlib to offer scalable workflows even for massive datasets.
Python shines in machine learning and AI applications in omics. Tools like scikit-learn, XGBoost, TensorFlow, and PyTorch let researchers build models to predict disease phenotypes, identify biomarkers, or classify subtypes of cancer.
✅ Learn Python step-by-step for bioinformatics with LBRN’s dedicated course:
Python for Omics Data Analysis – LBRN Training Platform
Timeline: Evolution of R and Python in Bioinformatics
| Year | Milestone |
|---|---|
| 2001 | Bioconductor launched for R, focused on genomics tools |
| 2008 | Biopython becomes stable for core sequence operations |
| 2015 | Seurat (R) and Scanpy (Python) emerge for single-cell analysis |
| 2020 | Rise of multi-omics integration and AI pipelines using Python |
| 2023+ | Cross-platform interoperability using reticulate and rpy2 gains traction |
Use Case Snapshots: What Can You Analyze?
Let’s briefly see the types of omics analyses possible with each language:
Transcriptomics (RNA-seq):
-
Python:
Scanpy,pandas,matplotlib,scikit-learn
Genomics (Variant Calling, SNPs):
Proteomics:
Metagenomics:
-
R:
phyloseq,microbiome -
Python:
QIIME 2,scikit-bio,biom-format
Epigenomics:
Multi-omics Integration:
R vs. Python: Which Should You Learn First?
-
Choose R if you’re interested in statistical genomics, gene expression, and custom data visualization.
-
Choose Python if you want pipeline automation, machine learning, or to build scalable web or cloud-based bioinformatics tools.
-
Best of both worlds? Learn both! In fact, many researchers use R for analysis and Python for automation or advanced modeling.
With packages like reticulate in R and rpy2 in Python, both languages can be used in a single notebook or pipeline.
Conclusion: Coding Is the New Microscope
In modern biology, coding is not just a technical skill-it’s the gateway to discovery. With R and Python, biologists can uncover hidden patterns in vast datasets, design better experiments, and even predict disease outcomes. These languages democratize data science, making it accessible, flexible, and reproducible for researchers at all levels.
If you're a student aiming to step into the world of omics, there’s no better time to start coding. The LBRN Training Platform offers free, beginner-friendly courses that not only teach the syntax but also contextualize coding in real bioinformatics research.
So whether you’re clustering single-cell data or mining genome variants, remember: the language of life is data-and R and Python are your translators.